GGMolVis Installation Guide with Blender
This guide provides instructions for installing GGMolVis and its dependencies, enabling you to use GGMolVis directly within Blender.
Installation Steps
Install Blender
Download and install Blender 4.3 or later from the official Blender website.
Install MolecularNodes
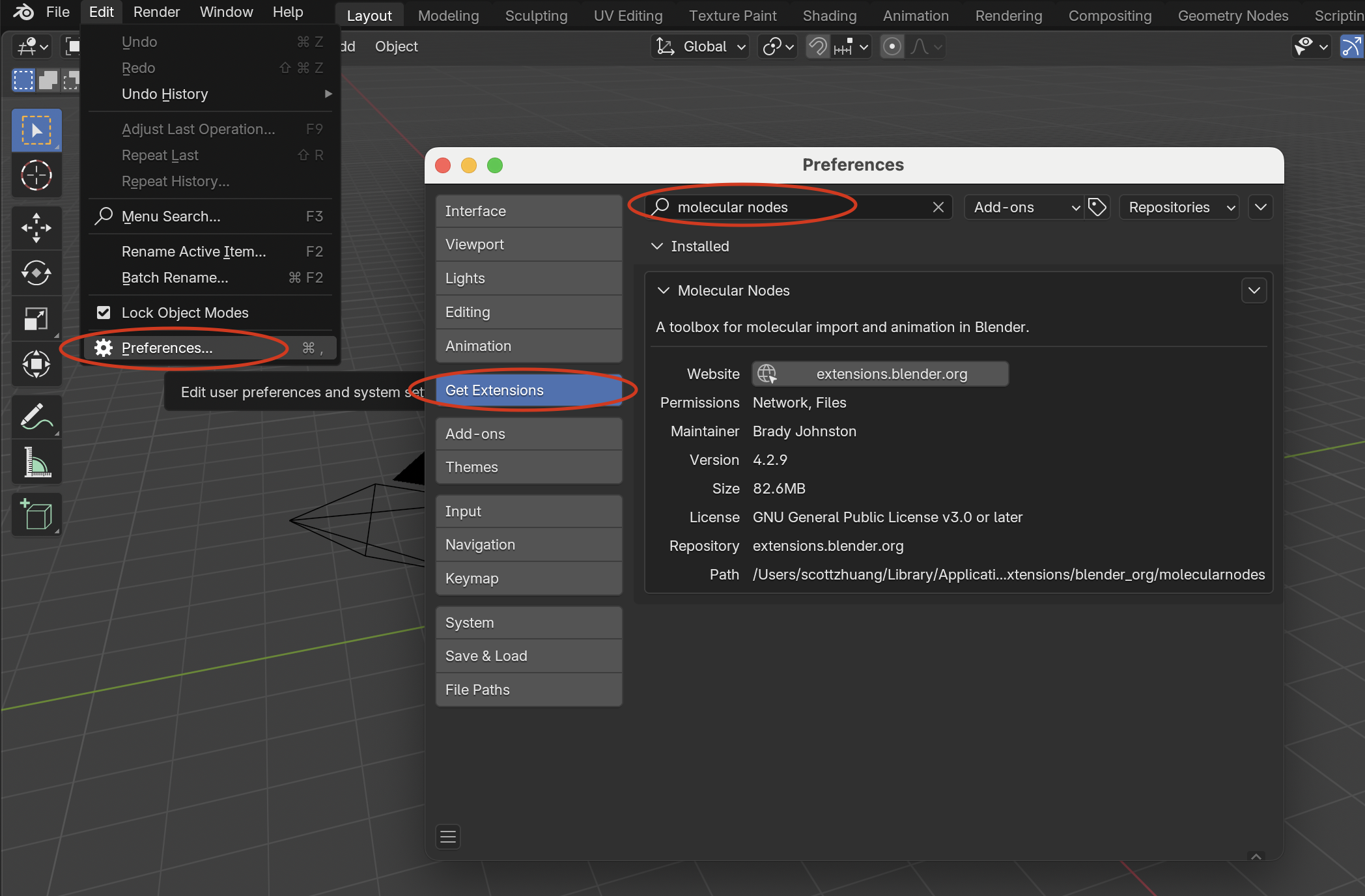
Within Blender, open Edit > Preferences:
Navigate to Get Extensions.
Search for Molecular Nodes and install it directly from the extensions list.
Install BNotebooks to Enable the Blender Jupyter Kernel
Download BNotebooks from the BNotebooks releases page.
Click the BNotebooks_0.0.5.zip link to download.
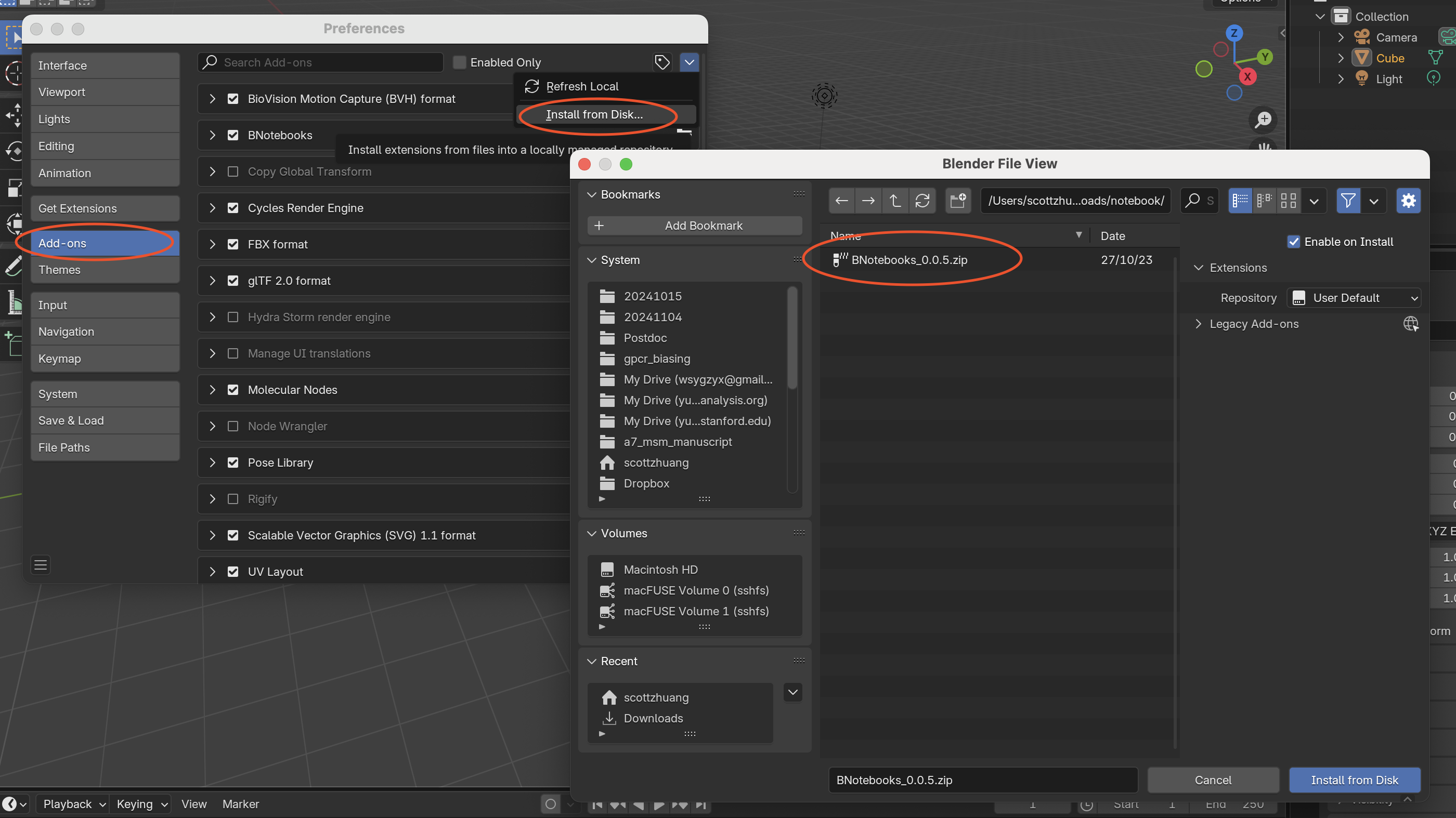
In Blender, go to Edit > Preferences > Add-ons:
Click the v button and select Install from Disk…
Navigate to the downloaded BNotebooks_0.0.5.zip file and click Install from Disk.
Enable the BNotebooks add-on by checking its box.
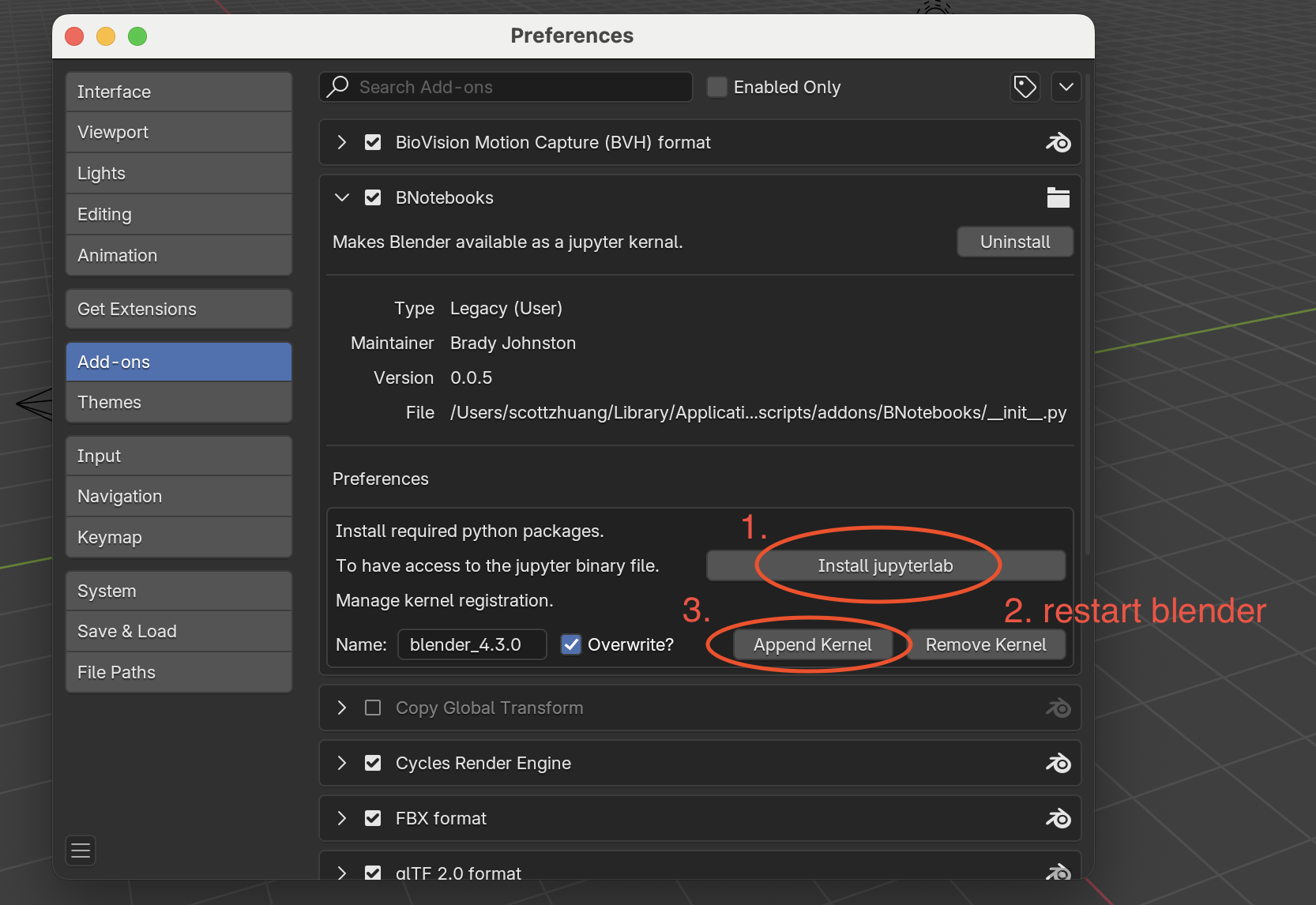
Click Install jupyterlab to install the JupyterLab extension.
Restart Blender to complete the installation.
After restarting Blender, return to Edit > Preferences > Add-ons and click Append Kernel to add the Jupyter kernel to Blender.
Install GGMolVis
Clone the GGMolVis repository:
git clone git@github.com:yuxuanzhuang/ggmolvis.git cd ggmolvis
Install GGMolVis using Blender’s bundled Python. Use the appropriate command for your operating system.
Note
You may need to adjust the path to the Python executable depending on where Blender is installed on your system.
macOS:
/Applications/Blender.app/Contents/Resources/4.3/python/bin/python3.11 -m pip install -e .Windows:
"C:\Program Files\Blender Foundation\Blender 4.3\4.3\python\bin\python3.11" -m pip install -e .Linux:
/usr/share/blender/4.3/python/bin/python3.11 -m pip install -e .
(Optional) Install a Different Version of MolecularNodes
If you need a development or alternative version of MolecularNodes:
git clone git@github.com:BradyAJohnston/MolecularNodes.git cd MolecularNodes /Applications/Blender.app/Contents/Resources/4.3/python/bin/python3.11 -m pip install -e .
Adjust the path as needed for your operating system.
Verification
Start a JupyterLab session:
jupyter labIn JupyterLab, select the blender_4.3.0 kernel. You should see a Blender application window open and connect to the Jupyter kernel.

In a Jupyter notebook cell, run the following commands to verify that GGMolVis is installed correctly:
import ggmolvis print(ggmolvis.__version__) print(ggmolvis.__file__)
If the commands produce the expected version number and file path, GGMolVis is successfully installed. You are now ready to use GGMolVis within Blender!